Last Updated on May 18, 2025 by Muhamed Elmesery
Before talking about cell cycle regulation, we should know more about the term “Cell Cycle” and what it means.
Table of Contents
Definition of Cell Cycle
Cell cycle is the process through which cells replicate and make two new cells.
We can also define cell cycle as a series of events that takes place in a cell as it grows and divides.
The cell cycle is generally divided into two phases: interphase and mitosis. During interphase, the cell spends most of its time performing the functions that make it unique. Mitosis is the phase of the cell cycle during which the cell divides into two daughter cells.
Stages of Cell Cycle
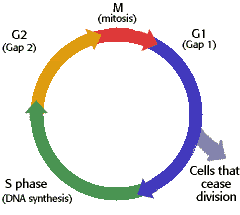
A cell spends most of its time in what is called interphase, and during this time it grows, replicates its chromosomes, and prepares for cell division. The cell then leaves interphase, undergoes mitosis, and completes its division. The resulting cells, known as daughter cells, each enter their own interphase and begin a new round of the cell cycle.
Interphase Stage
The interphase stage of the cell cycle includes three distinctive parts: the G1 phase, the S phase, and the G2 phase.
G1 is the stage where the cell is preparing to divide. To do this, it then moves into the S phase where the cell copies all the DNA. So, S stands for DNA synthesis. After the DNA is copied and there’s a complete extra set of all the genetic material, the cell moves into the G2 stage, where it organizes and condenses the genetic material, or starts to condense the genetic material, and prepares to divide.
Create a FREE Virtual Labs Account Now!
Mitosis Stage
The next stage is M. M stands for mitosis. This is where the cell actually partitions the two copies of the genetic material into the two daughter cells. After M phase completes, cell division occurs and two cells are left, and the cell cycle can begin again.
Mitosis is a continuous process, but for convenience in denoting which portion of the process is taking place, scientists divide mitosis into a series of phases:-
(1) Karyokinesis (Nuclear division) – divided into 4 sub-phases:-
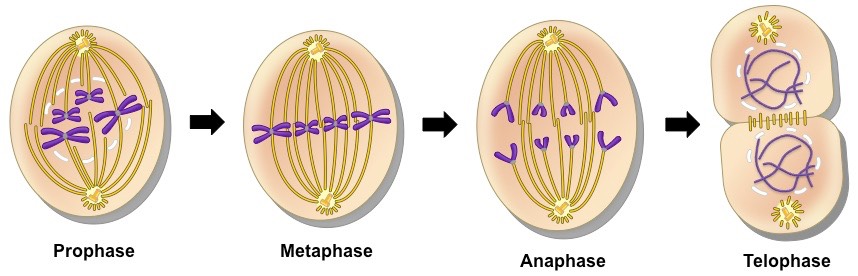
(a) Prophase
Mitosis begins with the condensing of the chromatin to form chromosomes in the phase called prophase. Two copies of each chromosome exist; each one is a chromatid. Two chromatids are joined to one another at a region called the centromere. As prophase unfolds, the chromatids become visible in pairs (called sister chromatids), the spindle fibers form, the nucleoli disappear, and the nuclear envelope dissolves.
In animal cells during prophase, microscopic bodies called centrioles begin to migrate to opposite sides of the cell. When the centrioles reach the poles of the cell, they produce and are then surrounded by a series of radiating microtubules called an aster. Centrioles and asters are not present in most plant or fungal cells.
As prophase continues, the chromatids attach to spindle fibers that extend out from opposite poles of the cell. The spindle fibers attach at the region of the centromere at a structure called the kinetochore, an area of protein in the centromere region. Eventually, all pairs of chromatids reach the center of the cell, a region called the equatorial plate.
(b) Metaphase
Metaphase is the stage of mitosis in which the pairs of chromatids line up on the equatorial plate. This region is also called the metaphase plate. In a human cell, 92 chromosomes in 46 pairs align at the equatorial plate. Each pair is connected at the centromere, where the spindle fiber is attached (more specifically at the kinetochore).
(c) Anaphase
At the beginning of anaphase, the sister chromatids move apart from one another. The chromatids are called chromosomes after the separation. Each chromosome is attached to a spindle fiber, and the members of each chromosome pair are drawn to opposite poles of the cell by the spindle fibers. During anaphase, the chromosomes can be seen moving. They take on a rough V shape because of their midregion attachment to the spindle fibers. The movement toward the poles is accomplished by several mechanisms, such as an elongation of the spindle fibers, which results in pushing the poles apart.
The result of anaphase is an equal separation and distribution of the chromosomes. In human cells, a total of 46 chromosomes move to each pole as the process of mitosis continues.
Get Started Praxilabs For FREE
(d) Telophase
In telophase, the chromosomes finally arrive at the opposite poles of the cell. The distinct chromosomes begin to fade from sight as masses of chromatin are formed again. The events of telophase are essentially the reverse of those in prophase. The spindle is dismantled and its amino acids are recycled, the nucleoli reappear, and the nuclear envelope is reformed.
(2) Cytokinesis (Division of Cytoplasm)
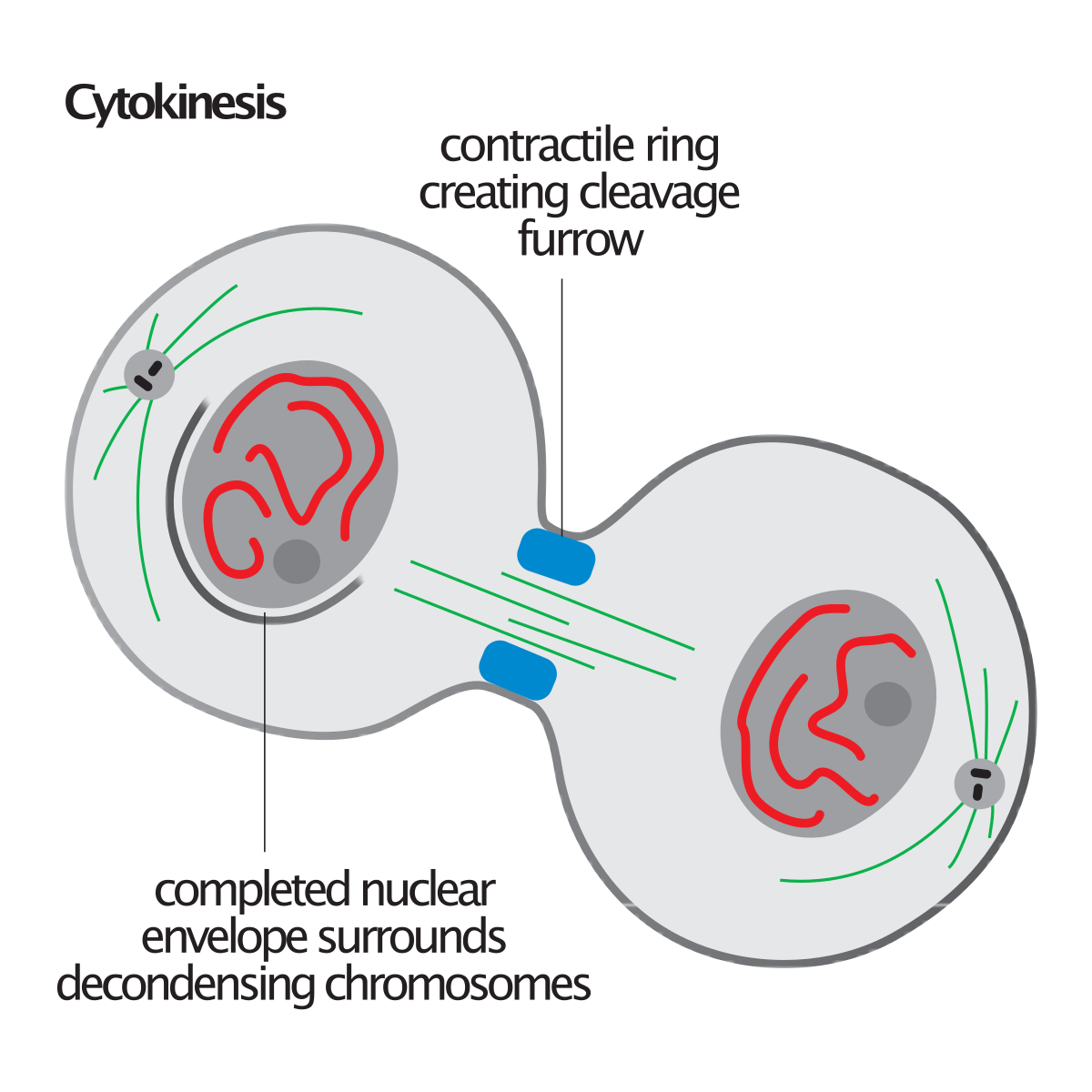
Leading to halving of that genome, passing into daughter cells.
Cytokinesis is the process in which the cytoplasm divides and two separate cells form. Note that cytokinesis is separate from the four stages of mitosis. In animal cells, cytokinesis begins with the formation of a cleavage furrow in the center of the cell. With the formation of the furrow, the cell membrane begins to pinch into the cytoplasm, and the formation of two cells begins. This process is often referred to as cell cleavage. Microfilaments contract during cleavage and assist the division of the cell into two daughter cells.
In plant cells, cytokinesis occurs by a different process because a rigid cell wall is involved. Cleavage does not take place in plant cells. Rather, a new cell wall is assembled at the center of the cell, beginning with vesicles formed from the Golgi apparatus. As the vesicles join, they form a double membrane called the cell plate. The cell plate forms in the middle of the cytoplasm and grows outward to fuse with the cell membrane. The cell plate separates the two daughter cells. As cell wall material is laid down, the two cells move apart from one another to yield two new daughter cells.
Mitosis serves several functions in living cells. In many simple organisms, it is the method for asexual reproduction (for example, in the cells of a fungus). In multicellular organisms, mitosis allows the entire organism to grow by forming new cells and replacing older cells. In certain species, mitosis is used to heal wounds or regenerate body parts. It is the universal process for cell division in eukaryotic cells.
We can summarized the functions of each phase as following:
G1 phase
- Cell increases in size.
- Cellular contents are duplicated.
S phase
- DNA replication.
- Each of the 46 chromosomes (23 pairs) is replicated by the cell.
G2 phase
- Cell grows more.
- Organelles and proteins develop in preparation for cell division.
M phase
- Mitosis followed by cytokinesis (cell separation).
- Formation of two identical daughter cells.
G0 phase
While some cells are constantly dividing, some cell types are quiescent. These cells exit G1 and enter a resting state called G0. In G0, a cell is performing its function without actively preparing to divide. G0 is a permanent state for some cells, while others may restart division if they get the right signals.
Regulation of Cell Cycle (cyclins and cdks)
How is the cell cycle regulated?
Cell cycle does not occur in unchecked manner. The preparations of the cell are checked by regulatory molecules which are proteins that are responsible about cell cycle regulation . It includes the detection and repair of genetic damage as well as prevention of uncontrolled cell division. There are two key classes of regulatory molecules (cell cycle regulators) that determine the mechanism of cell cycle regulation :
► Cyclins
► Cyclin dependent kinases (Cdk)
Cyclins
What do cyclins do? (Role of cyclins in cell cycle regulation)
Cyclins Determine the Activity of CDKs (named because their levels change during the cell cycle).
Cyclins are divided into four classes defined by their presence and activity during the cell cycle:
- G1 Cyclins (D cyclins)
- G1/S cyclin (Cyclin E)
- S-phase cyclins (cyclins E and A)
- M-phase cyclins (B cyclins)
-
G1 cyclins (Cyclin D)
coordinates the cell cycle with extracellular events. Their activity is subject to regulation by signal transduction pathways that sense the presence of growth factors or cell proliferation inhibitory signals. G1 cyclins (cyclin D) interact with CDK4 and CDK6 and promote the entry into the cell cycle.
-
G1/S cyclin (Cyclin E)
The G1/S cyclins accumulate during late G1, reach peak levels when cells enter S phase, and decline during S phase. They are known as cyclin E and bind to CDK 2. The main function for the cyclinE -CDK 2 complex, together with cyclinD-CDK4/6, is to trigger the G1-S phase transition. This transition is known as START and is defined as the point at which cells are irreversibly committed to cell division and can no longer return to the G1 state.
- S phase cyclins (Cyclin A and Cyclin E)
S phase cyclins are synthesized at the end of G1, levels remain high throughout the S phase and do not decline until early mitosis. Two types of S phase cyclins trigger S phase in: cyclin E, which can also promote entry into the cell cycle and is therefore also a G 1/S cyclin, and cyclin A. Both cyclins bind CDK 2 and are directly responsible for DNA synthesis.
Try DNA synthesis in Praxilabs Virtual Labs
-
Mitotic cyclins
Mitotic cyclins bind CDK1 to promote entry into and progression through mitosis. The mitotic cyclins are cyclin A and cyclin B Mitotic cyclin-CDK complexes are synthesized during S phase and G2, but their activities are held in check until DNA synthesis is completed.
The different types of cyclins are quite distinct from each other in protein sequence, but all of them contain a conserved region known as the cyclin box and possess similar three-dimensional structures three key features of Cyclins:
- Cyclins bind to and activate CDKs. The activity and substrate specificity of any given CDK is primarily defined by the particular cyclin to which it is bound.
- Cyclins are only present during the cell cycle stage that they trigger and are absent in other cell cycle stages.
- Cyclins not only regulate a particular cell cycle stage but also set in motion a series of events in preparation for the next cell cycle stage. In this way, they propel the cell cycle forward.
Cyclin Expression Cycle
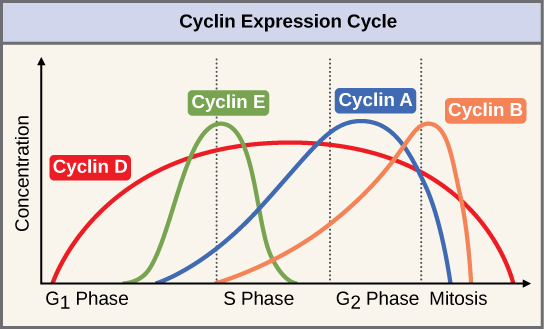
Once activated, mitotic CDKs promote entry into mitosis by phosphorylating and activating hundreds of proteins to promote chromosome segregation and other aspects of mitosis. Their inactivation during anaphase prompts cells to exit mitosis, which involves the disassembly of the mitotic spindle, chromosome, the re-formation of the nuclear envelope, and eventually cytokinesis.
Cyclin dependent kinases (cdks cell cycle )
Cyclin-dependent kinases (CDKs) are protein kinases characterized by needing a separate subunit – a cyclin – that provides domains essential for enzymatic activity.
CDKs play important roles in cell cycle regulation( the control of cell division and modulate transcription in response to several extra- and intracellular cues). The evolutionary expansion of the CDK family in mammals led to the division of CDKs into three cell-cycle-related subfamilies :
G1 Cdk (Cdk 4)
S-phase Cdk (Cdk 2)
M-phase Cdk (Cdk 1)
► Their levels in the cell remain stable.
► Remain inactive.
► Bind to the appropriate cyclin in order to be activated.
►Their function is to provide phosphate gr to a no. of proteins that control processes in the cell cycle.
A CDK binds a regulatory protein (cyclin). Without cyclin, CDK has little kinase activity; only the cyclin-CDK complex is an active kinase but its activity can be typically further modulated by phosphorylation and other binding proteins, like p27.
Cyclin CDKs Complex
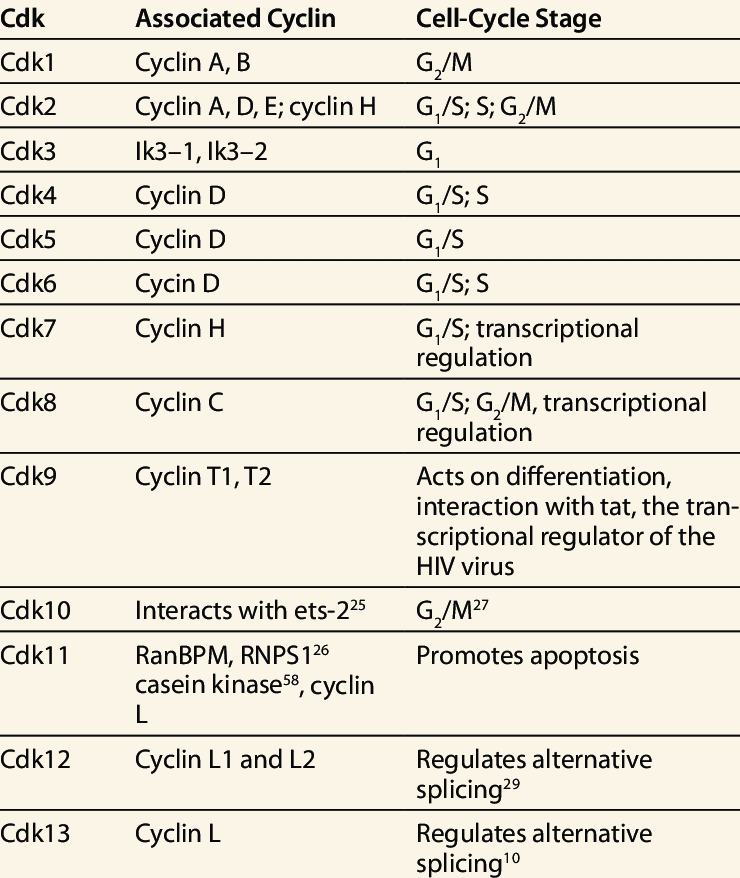
The previous table shows cyclin – cyclin dependent kinases (Cdk) complexes formed during cell cycle regulation and their functions
Regulation of CDK Activity
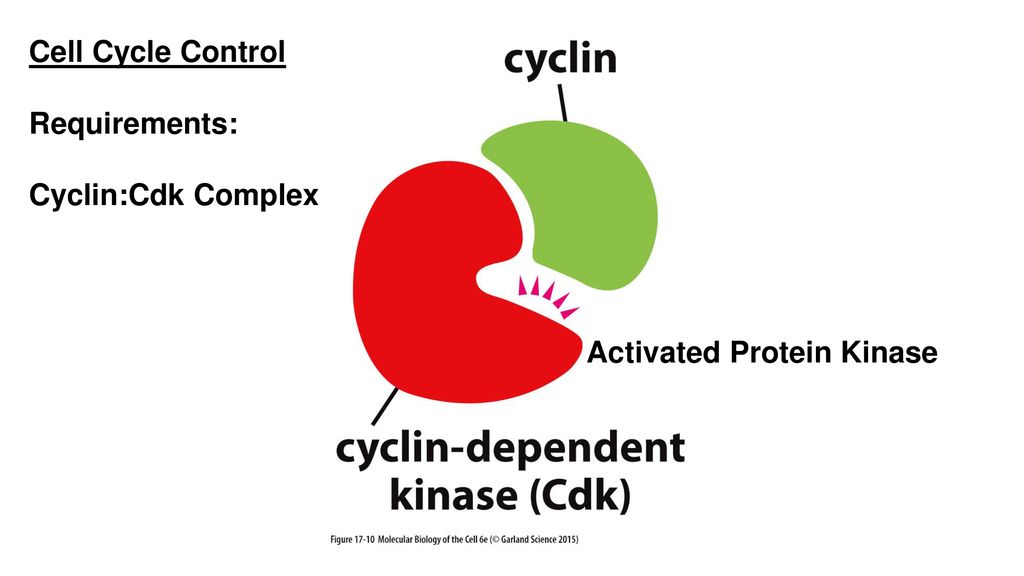
Mechanism of cell cycle regulation by CDKs activation
Multiple mechanisms ensure that CDKs are active in the right stage of the cell cycle. CDK activity is regulated by multiple mechanisms:
- Regulating the cyclin levels
- Action of CDK-activating kinase (CAK)
- Inhibitory phosphorylations on CDK
- Action of CDK Inhibitors
- Regulation of Cyclin Levels
The timely activation of CDKs depends, in part, on the presence of the appropriate cyclins in the cell cycle stage where they are needed.
Cells utilize multiple mechanisms to restrict cyclins to the appropriate cell cycle stage and to keep them at the right concentration.
1.Regulation of Cyclin Mechanisms:
- Transcriptional control of cyclin genes.
- Degradation of cyclins.
- Transcriptional control of the cyclin subunits is one mechanism that ensures proper temporal expression of the cyclins.
- Degradation of cyclins.
The most important regulatory control that restricts cyclins to the appropriate cell cycle stage is ubiquitin-mediated protein degradation.
Cyclins are degraded through the action of two different ubiquitin-proteins:
- a) SCF (Skp1,Cullin & f-box proteins)
- b) APC/C (anaphase-promoting complex or cydosome)
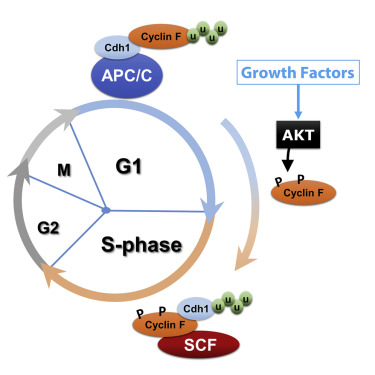
SCF controls the G1-S phase transition by degrading G1 cyclins (Cyclin D) The APC/C degrades S phase and mitotic cyclins, thereby promoting the exit from mitosis.
Regulating the levels of cyclins is not the only mechanism that controls CDK activity.
Activating and inhibitory phosphorylation events on the CDK subunit and presence of inhibitors are essential to the control cyclin-CDK activity.
- CDK-activating kinase (CAK)
Phosphorylation of a threonine residue is required for CDK activity. This phosphorylation is mediated by the CDK-activating kinase (CAK). CAK activity is constant throughout the cell cycle and phosphorylates the CDK as soon as a cyclin-CDK complex is formed.
- Inhibitory phosphorylations on CDK
Inhibitory phosphorylation on CDK plays a critical role in controlling CDK activity. A kinase called “Weel” brings about this inhibitory phosphorylation.
- CDK Inhibitors (CKIs)
CDK inhibitors control Cyclin-CDK activity
Family of proteins known as CDK inhibitors or CKIs, that directly bind to the cyclin-CDK complex and inhibit its
activity.
These proteins play an especially important role in the regulation of the G1-S phase transition (entry into the cell cycle).the genes encoding these CKIs are often found mutated in human cancers CKIs involved in regulating S phase and mitotic CDKs are all essential to prevent premature activation of S phase and M phase CDKs.
Inhibitors of G1 CDKs play an essential role in mediating a G1 arrest in response to proliferation inhibitory signals.
A class of CKIs called INK4s interact only with the G1 CDKs. Binding of INK4s to CDK4 and CDK6 blocks their interaction with cyclin D and hence their protein kinase activity.
A second class of CKIs found in metazoan cells consists of three proteins; p21 p27 and p57.
p53 is a TF, which stimulates the expression of p21, p27 and p57 genes.
The p21 protein inhibits cyclin/CDK protein complexes that are needed to progress from the G1 phase of the cell cycle to the S phase. CKIs regulating G1 CDKs play a critical role in preventing tumor formation.
CDK Inhibitors
One of important cyclins that play a vital role in cell cycle regulation is cyclin-dependent kinase inhibitor (CKI) is a protein that interacts with a cyclin-CDK complex to block kinase activity, usually during G1 or in response to signals from the environment or from damaged DNA. In animal cells, there are two major CKI families: the INK4 family and the CIP/KIP family. The INK4 family proteins are strictly inhibitory and bind CDK monomers.
Crystal structures of CDK6-INK4 complexes show that INK4 binding twists the CDK to distort cyclin binding and kinase activity. The CIP/KIP family proteins bind both the cyclin and the CDK of a complex and can be inhibitory or activating.
Cell Cycle Checkpoints
Cell cycle checkpoints are used by the cell to monitor and regulate the progress of the cell cycle. The cell can’t proceed to the next phase until checkpoint requirements have been met.
There are three main checkpoints:
1) G1/S checkpoint (before cell enters S phase).
2) G2/M checkpoint (after S phase).
3) APC/C checkpoint (during mitosis).
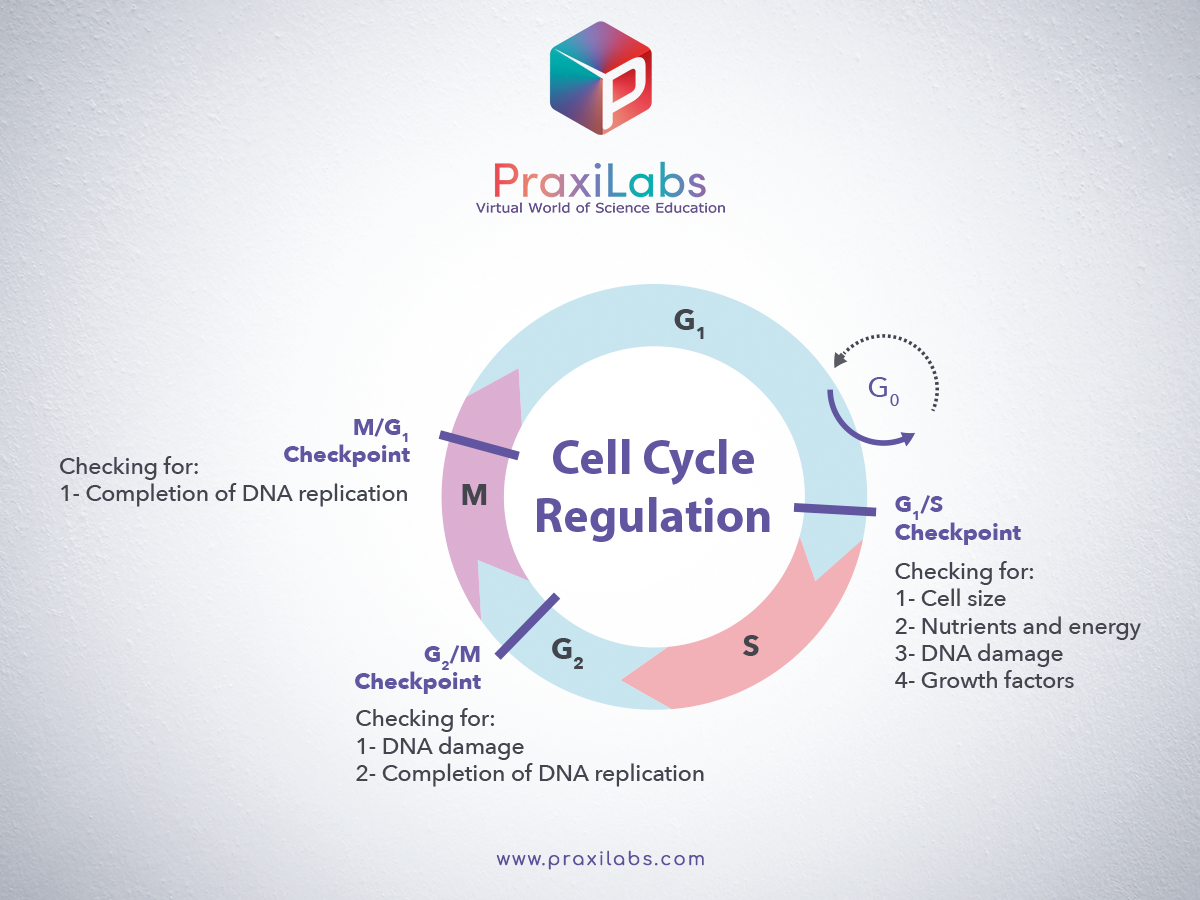
1) G1/S checkpoint (before cell enters S phase):
► Checks for cell size
► Checks for nutrients
► Checks for DNA damage
► Checks for all the preparations (all proteins, ATP etc. requires in S phase)
► Checks whether S phase Cyclins and Cdk complex is activated to initiate DNA replication.
Then the cell passes to the next S phase.
2) G2/M checkpoint (after S phase):
► Checks for proper DNA replication
► Checks for all the preparations (all proteins, ATP etc. required in M phase)
► Checks for Tubulin synthesis
► Checks whether M phase Cyclins and Cdk complex is activated to initiate mitosis
Then the cell passes to the next M phase.
All the checkpoints require the services of a complex of proteins. The levels of these proteins are increased in damaged cells. They allow time to prepair DNA by blocking the cell cycle.
P53 is a protein which senses DNA damage and can halt progression of the cell cycle in G1 phase by blocking the activity of Cdk2 until damage can be repaired. If the damage is so severe that it can’t be repaired, then the cell destructs itself by apoptosis. In case of damage to DNA after S phase, the action of Cdk1 is inhibited, thus stopping progression of the cell from G2 to mitosis.
Quick Facts about the Cell Cycle
- Leland H. Hartwell, Tim Hunt and Sir Paul M. Nurse were jointly awarded the Nobel Prize in Physiology or Medicine 2001 for discovering the cell cycle key regulators.
- There is more DNA in the S phase cells than that exists in the cells of the G1 phase. In G2, the cells will approximately have twice the DNA than that exists in G1. Therefore, more dye will be consumed in the G2. Cells’ suspension is then aspirated into a flow cell. Surrounded by a narrow stream of the fluid, the cells will pass through a focused laser beam. When the light strikes a cell, it is either absorbed or scattered.
- If the absorbed light is of an appropriate wavelength, thus it is re-emitted as fluorescence. This process shows and reflects the cell’s size and shape, i.e. the cell’s internal structure. By using a computer system and throughout a series of photodiodes, the scattering of the fluorescence signals is detected, amplified and then analyzed.
- Quantitative information about every analyzed cell is the end result of this whole process. In a short period of time, an enormous number of cells is analyzed (>1000 cells/second). This verifies the creation of statistically well-founded information about cell populations.
Try Our Virtual Lab of Experiment Flow Cytometry- Cell Cycle
Flow cytometry is a laser-based technology that allows quantitative single cell analysis. It is used to analyze the characteristics of cells. Analysis of the Cell cycle by DNA quantification can be done using flow cytometry.
Cells are fixed and permeabilized using ethanol. Then, propidium iodide (PI) is used for staining of DNA in intact cells. RNase is used to ensure only DNA is stained with PI.unstained cells are used as control.
Our virtual laboratory of Flow Cytometry allows the students to practice the steps of cell fixation and permeabilization and understand the concept of cell cycle analysis using propidium iodide.
Create your free account and try our simulations
 PraxiLabs A virtual world of science
PraxiLabs A virtual world of science





